Useful error messages.... and another format rant.
I sent a query to Biomart the other day, as I often do. Most of the time, I get back my results quickly, and have no problems whatsoever. It's one of my "go-to" sites for useful genomic data. Unfortunately, every time I tried to download the results of my query, I'd get 2-3Mb into the file before the download would die. (It was a LONG list of snps, and the file size was supposed to be in the 10Mb ballpark.)
Anyhow, in frustration, I tried the "email results to you" option, whereupon I got the following email message:
Your results file FAILED.
Here is the reason why:
Error during query execution: Server shutdown in progress
That has to be the first time I've ever had a server shutdown cause a result failure. Ok, it's not that funny, but I am left wondering if that was the cause of the other 10 or so aborted downloads. Anyone know if Biomart runs on Microsoft products? (-;
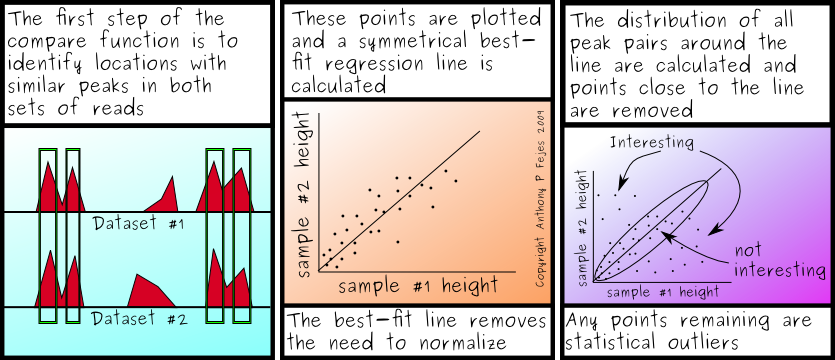
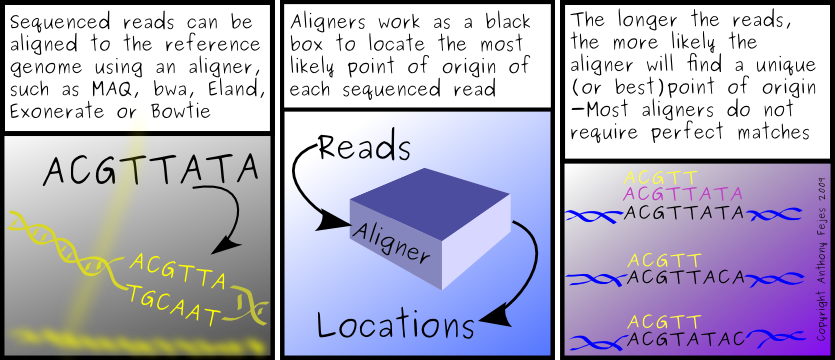
The other thing on my mind this afternoon is that I am still looking to see my first Variant Call Format file for SNPs. A while back, I was optimistic about seeing the VCF files in the real world. Not that I can complain, but I thought adoption would be a little faster. A uniform SNP format would make my life much more enjoyable - I now have 7 different SNP format iterators to maintain, and would love to drop most of them.
What surprised me, upon further investigation, is that I'm also unable to find a utility that actually creates VCF files from .map, SAM/BAM, eland, bowtie or even pileup files. I know of only one SNP caller that creates VCF compatible files, and unfortunately, it's not freely available, which is somewhat un-helpful. (I don't know when or if it will be available, although I've heard rumours about it being put into our pipeline...)
That's kind of a sad state of affairs - although I really shouldn't complain. I have more than enough work on my plate, and I'm sure the same can be said for those who are actively maintaining SNP callers.
In the meantime, I'll just have to sit here and be patient... and maybe write an 8th snp format iterator.
Labels: Bioinformatics, documentation, formats, SNP calling, SNP Database, SNPS, Software