Base quality by position
A colleague of mine was working on a nifty tool to give graphs of the base quality at each position in a read using Eland Export files, which could be incorporated into his pipeline. Over a discussion about the length of time it should take to do that analysis, (His script was taking an hour, and I said it should take about a minute to analyze 8M illumina reads...) I ended up saying I'd write my own version to do the analysis, just to show how quickly it could be done.
Well, I was wrong about it taking about a minute. It turns out that the file has a lot more than about double the originally quoted 8 million reads (QC, no match and multi match reads were not previously filtered), and the whole file was bzipped, which adds to the processing time.
Fortunately, I didn't have to add bzip support in to the reader, as tcezard (Tim) had already added in a cool "PIPE" option for piping in whatever data format I want in to applications of the Vancouver Short Read Analysis Package, thus, I was able to do the following:
Quite a neat use of piping, really.
Anyhow, the fun part is that this was that the library was a 100-mer illumina run, and it makes a pretty picture. Slapping the output into openoffice yields the following graph:
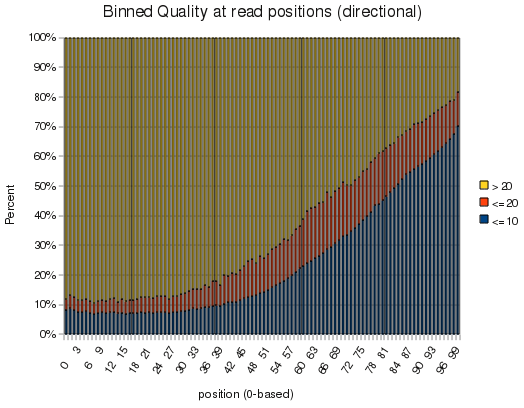
I didn't realize quality dropped so dramatically at 100bp - although I remember when qualities looked like that for 32bp reads...
Anyhow, I'll include this tool in Findpeaks 4.0.8 in case any one is interested in it. And for the record, this run took 10 minutes, of which about 4 were taken up by bzcat. Of the 16.7M reads in the file, only 1.5M were aligned, probably due to the poor quality out beyond 60-70bp.
Well, I was wrong about it taking about a minute. It turns out that the file has a lot more than about double the originally quoted 8 million reads (QC, no match and multi match reads were not previously filtered), and the whole file was bzipped, which adds to the processing time.
Fortunately, I didn't have to add bzip support in to the reader, as tcezard (Tim) had already added in a cool "PIPE" option for piping in whatever data format I want in to applications of the Vancouver Short Read Analysis Package, thus, I was able to do the following:
time bzcat /archive/solexa1_4/analysis2/HS1406/42E6FAAXX_7/42E6FAAXX_7_2_export.txt.bz2 | java6 src/projects/maq_utilities/QualityReport -input PIPE -output /projects/afejes/temp -aligner elandext
Quite a neat use of piping, really.
Anyhow, the fun part is that this was that the library was a 100-mer illumina run, and it makes a pretty picture. Slapping the output into openoffice yields the following graph:

I didn't realize quality dropped so dramatically at 100bp - although I remember when qualities looked like that for 32bp reads...
Anyhow, I'll include this tool in Findpeaks 4.0.8 in case any one is interested in it. And for the record, this run took 10 minutes, of which about 4 were taken up by bzcat. Of the 16.7M reads in the file, only 1.5M were aligned, probably due to the poor quality out beyond 60-70bp.
Labels: application development, Open Source, programming, Sequencing, Solexa/Illumina, Vancouver Short Read Analysis Package

2 Comments:
6 minutes per 8M reads is still very slow. I know a C program can generate similar stats in ~40 seconds given 8M 36bp reads (FASTQ format). 100bp should take about 2 minutes at this rate. I also think that script is not optimized. I am sure it will take longer, but should not be an hour. In addition, bzip2 is several times slower than gzip on decompression. That is why all big files are compressed by gzip instead of bzip2.
Excellent comment - except you're not comparing apples with apples.
(6 minutes for 8M reads is slow, I can't argue that.)
I should point out a few details I left out. This was ~6 minutes for 17M 100bp reads on an older machine (with many other users fighting for resources) with a network mounted drive.
The performance, obviously, increases dramatically if you run with 36bp instead of 100bp, since the only operation this particular app is doing is working on the per-base quality scores.
When I use the "PIPE" option on VSRAP applications, I lose a lot of performance gains built into the java file handlers.
And yes - bzip2 was FAR from my first choice of compression. (-;
Sometimes you just have to do what you have to do.
The only valid comparison that can be done are the two different programs (mine and my colleagues) that were run on the same machine.
For the record, on a more competitive, eg, not wildly overloaded, machine, I was getting performance closer to 8 seconds using a maq file with 700,000 36bp reads - which still comes out to just over a minute for 8M reads.
For whatever it's worth, java will probably never be as fast as C.... but there are gains in other aspects that make up for it. I'm willing to wait an extra 40 seconds for my results, but 40 minutes is way out of the question. (-:
Post a Comment
<< Home