Another day, another result...
If you follow along with Daniel McArthur's blog, Genetic Future, you may have caught the announcement that Illumina is getting into the personal genome sequencing game. While I can't admit that I was surprised by the news, I will have to admit that I am somewhat skeptical about how it's going to play out.
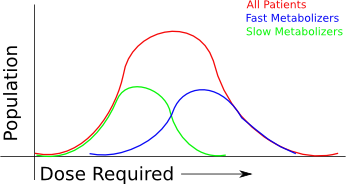
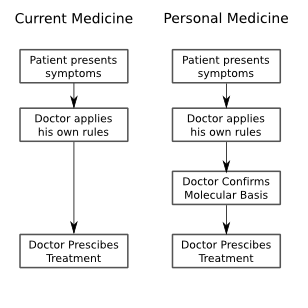
If your business is using arrays, then you'll have an easy time sorting through the relevance of the known "useful" changes to the genome - there are only a couple hundred or thousand that are relevant at the moment, and several hundred thousand more that might be relevant in the near future. However, when you're sequencing a whole genome, interpretation becomes a lot more difficult.
Since my graduate project is really the analysis of transcriptome sequencing (a subset of genome sequencing), I know firsthand the frustration involved. Indeed, my project was originally focused on identifying changes to the genome common to several cancer cell lines. Unfortunately, this is what brought on my need to rant: there is vastly more going on in the genome than small sequence changes.
We tend to believe blindly what we were taught as the "central paradigm of molecular biology". Genes are copied to mRNA, mRNA is translated to proteins, and the protein goes off to do it's work. However, cells are infinitely more complex than that. Genes can be inactivated by small changes, can be chopped up and spliced together to become inactivated or even deregulated, interference can be run by distally modified sequences, gene splicing can be completely co-opted by inactivating genes we barely even understand yet and desperately over-expressed proteins can be marked for deletion by over-activating garbage collection systems so that they don't have a chance to get where they were needed in the first place. And here we are, looking for single nucleotide variations, which make up a VERY small portion of the information in a Cell.
I don't have the solution, yet, but whatever we do in the future, it's not going to involve $48,000 genome re-sequencing. That information on it's own is pretty useless - we'll have to study expression (WTSS or RNA-Seq, so figure another $30,000), changes to epigenetics (of which there are many histone marks, so figure 30 x $10,000) and even dna methylation (I don't begin to know what this process costs.)
So, yes, while I'm happy to see genome re-sequencing move beyond the confines of array based SNP testing, I'm pretty confident that this isn't the big step forward it might seem. The early adopters might enjoy having a pretty piece of paper that tells them something unique about their DNA, and I don't begrudge it. (In fact, I'd love to have my DNA sequenced, just for the sheer entertainment value.) Still, I don't think we're seeing a revolution in personal genomics - not quite yet. Various experiments have shown we're on the cusp of a major change, but this isn't the tipping point: we're still going to have to wait for real insight into the use of this information.
When Illumina offers a nice toolkit that allows you to get all of the SNVs, changes in expression and full ChIP-Seq analysis - and maybe even a few mutant transcription factor ChIP-Seq experiments thrown in - and all for $48,000, then we'll have a truly revolutionary system.
In the meantime, I think I'll hold out on buying my genome sequence. $48,000 would buy me a couple more weeks in Tahiti, which would currently offer me a LOT more peace of mind. (=
And on that note, I'd better get back to doing the things I do.... new FindPeaks tag, anyone?
Labels: Bioinformatics, Genomics, personalized medicine, rant, Solexa/Illumina